Since 2005 when the Imaging Response Assessment Team (IRAT) program was put into place at UPMC Hillman Cancer Center, significant infrastructure has been developed to facilitate collaborations between IRAT teams and clinical leaders. The ongoing success of this program requires that its function continue as a major component of the In Vivo Imaging Facility as the Image Protocol Development and Data Management (IPDDM) service. IRAT administrative mechanisms are now embedded into the protocol approval and research operations of UPMC Hillman Cancer Center to ensure interactions between Radiology Imaging Service Divisions and Disease Site Oncology Group Leaders are maintained. The IPDDM protocol administrator/clinical coordinator (with a central office located at UPMC Hillman Cancer Center) facilitates the incorporation of imaging into cancer therapy trials by modifying or writing new clinical protocols and ensuring they receive timely review through the institutional IRB committees.
As a result, a number of new quantitative imaging biomarker therapy response assessment trials have been initiated. Additionally, Dr. Mountz was awarded several related grants from the NIH, including the U01, “Early Cancer Therapy Response Assessment Using Quantitative Imaging Biomarkers,” investigating imaging biomarker response to therapies in glioblastoma multiforme and squamous cell carcinoma of the head and neck; and a high-end instrumentation grant, “PET/CT for Quantitative Biomarker Assessment of Early Disease and Therapy Response” to fund a dedicated research whole body, state-of-the-art PET/CT scanner. An example of imaging protocol design to assess early cancer therapy response is shown in Figure 1.

Figure 1. Example PET-CT scan timeline that could be used for imaging biomarkers in a cancer therapy drug trial.
The Tumor Imaging Metric (TIM) analysis laboratory, associated with the IPDDM, provides a growing array of consultation and analysis services to a growing customer base (Figure 2).
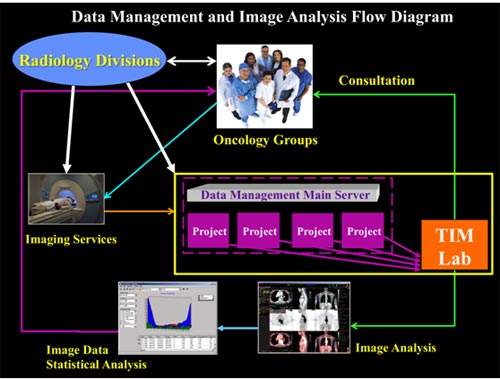
Figure 2. Project workflow through the Tumor Imaging Metric (TIM) Analysis Laboratory. The process shows the information and communication network. The Radiology Divisions interact with the Oncology Groups, the imaging staff, and the data analysis personnel. The Hillman/UPMC PET core has obtained space and purchased a dedicated oncology workstation (MIMVista, Cleveland, OH) and PACS (Stentor, ISOBAR) to allow for routine and efficient assessment of tumor volume and total tumor metabolic index. The laboratory has the capability of uploading and registration of the same patient over multiple time points, and then directly comparing image biomarker change at baseline, intervals during therapy, and follow-up. As the projects are underway, the TIM Laboratory personnel consult with the oncology groups to report feedback and assure cohesive work. Lastly, the completed quantitative image analysis and statistical data analysis are presented back to the oncology group leaders.
TIM Lab Image Analysis
Services offered include an assortment of simplified imaging biomarker procedures depending on the needs of the researcher (Figures 3 and 4). Examples include assessment of static tracer uptake measures such as SUV, maximum SUV, and tumor glycolytic index (in the case of FDG). PET-CT scans of one patient at different therapy time-points can be registered together in order to evaluate therapeutic response over time.

Figure 3. One arm of the TIM analysis laboratory is the development of simplified, clinically practical methods for evaluating early-therapy response in cancers. Here, a 60 year old patient with squamous cell carcinoma of the head and neck (SSCHN) is demonstrating a large 3.3×2.3cm confluent nodal metastasis at left level II. Volumetric method of analysis (left) is performed by drawing a volume of interest (VOI) around the lesion and mirrored contra-lateral region is displayed here in a series of transverse slices. The baseline scan (right, top row) maximum SUV was 17.7 and the post therapy (right, bottom row) maximum SUV was 3 were found with conventional clinical ROI measurements. Total glycolytic index (TGI) was also measured at both time points. At baseline, TGI= 82.2 and decreased to 4.6 at post-therapy, which was within one SD of background FDG activity.

Figure 4. SUV threshold defined region of interest precisely translated to CT imaging at both baseline and therapy response.
The TIM lab also offers more advanced image analysis methods, such as full kinetic modeling of dynamic PET image data (Figure 5). Additionally, simplified graphical (e.g., Logan, Patlak) and reference tissue methods can be performed in lieu of a full kinetic analysis.

Figure 5. Example results obtained during a dynamic PET study, which incorporates modeling of kinetic tracer uptake parameters. Left, shows a late time [18F]Fluorothymidine PET image of a brain tumor with region of interest (orange). Center, shows an image-based input function [(IBIF); blue], obtained from the internal carotid arteries, and a tumor activity curve [(TAC); red], obtained from a subject’s baseline dynamic PET scan. IBIF and TAC are used as compartmental modeling inputs (top, right) from which kinetic rate parameters can be obtained (bottom right).
A major accomplishment of IPDDM has been the continued growth in the number of oncology projects utilizing imaging for the development and evaluation of PET and MR biomarkers to assess efficacy of new cancer therapies in clinical trials. The establishment of state-of-the-art PET-CT and MR imaging, protocol development, and data analysis in oncology has resulted in steadily increasing collaborative interactions between Hillman investigator-initiated clinical trial personnel and members of the Department of Radiology at UPMC.


